
Recently, the 10th annual Laboratory of Clinical Microbiology (LCM) Poster Day was held at the David Axelrod Institute.
LCM Research Seminars take place monthly, September through June. This annual poster session kicks off the 31st year of seminars.
Wadsworth Center staff from multiple sites attended. Highlights of eleven posters, all previously presented at scientific meetings locally and nationwide, follow.
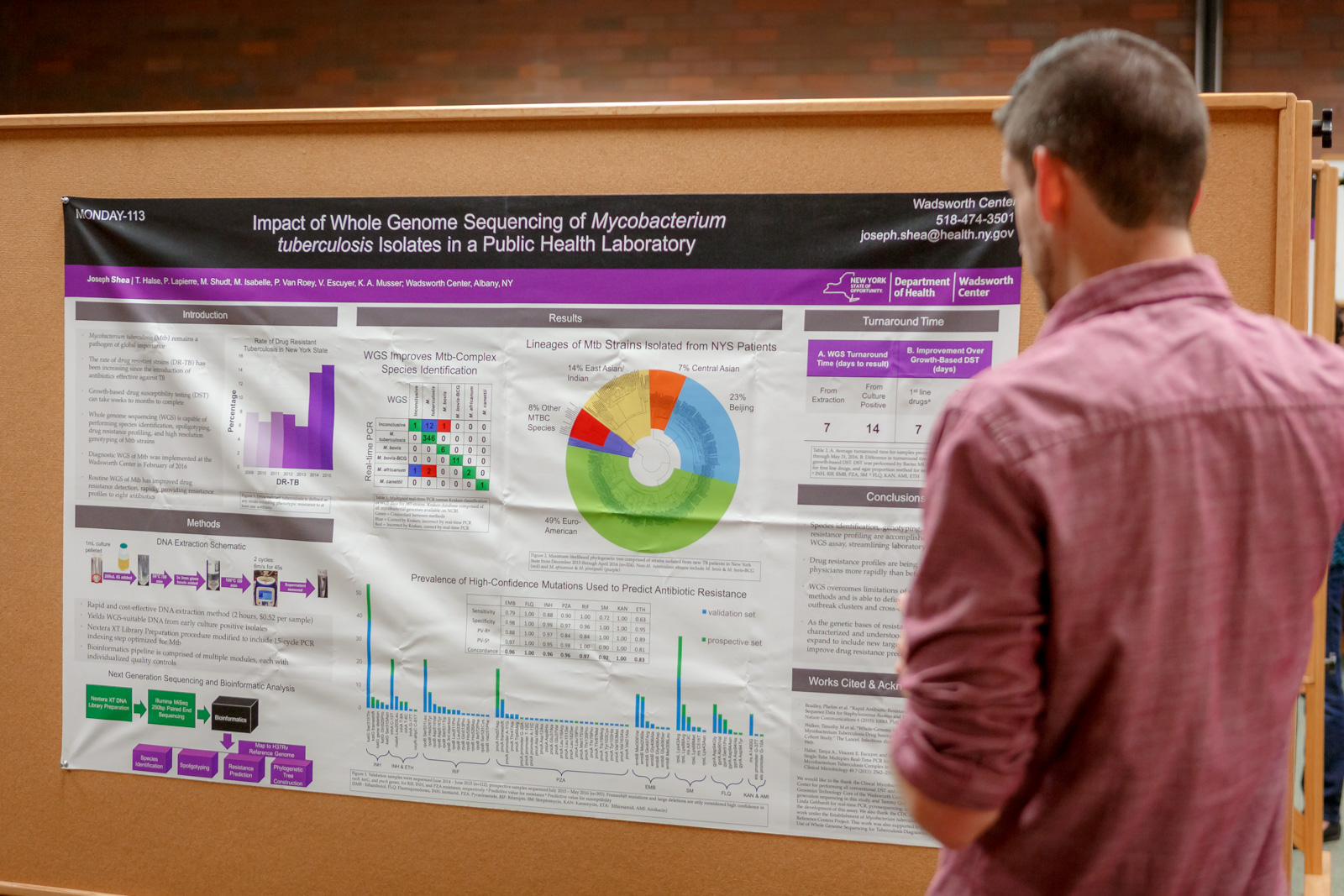 Whole genome sequencing (WGS) can replace multiple molecular tests, reducing turn-around-time as well as the cost of testing. Work is on-going in the Bacteriology Laboratory to obtain New York State approval to report serogroup and virulence data for Shiga toxin-producing E. coli.
Whole genome sequencing (WGS) can replace multiple molecular tests, reducing turn-around-time as well as the cost of testing. Work is on-going in the Bacteriology Laboratory to obtain New York State approval to report serogroup and virulence data for Shiga toxin-producing E. coli.Similarly, the Mycobacteriology Laboratory is using WGS of Mycobacterium tuberculosis in place of multiple molecular tests and has found they can accurately and more quickly determine the species, genotype and drug resistance profile. The laboratory has received NYS approval to report WGS test results.
Because Salmonella enterica serovar Enteritidis is not genetically diverse, there are occasions when specimens cannot be differentiated by pulse field gel electrophoresis. The Bacteriology Laboratory is studying the utility of WGS in surveillance and outbreak investigations.
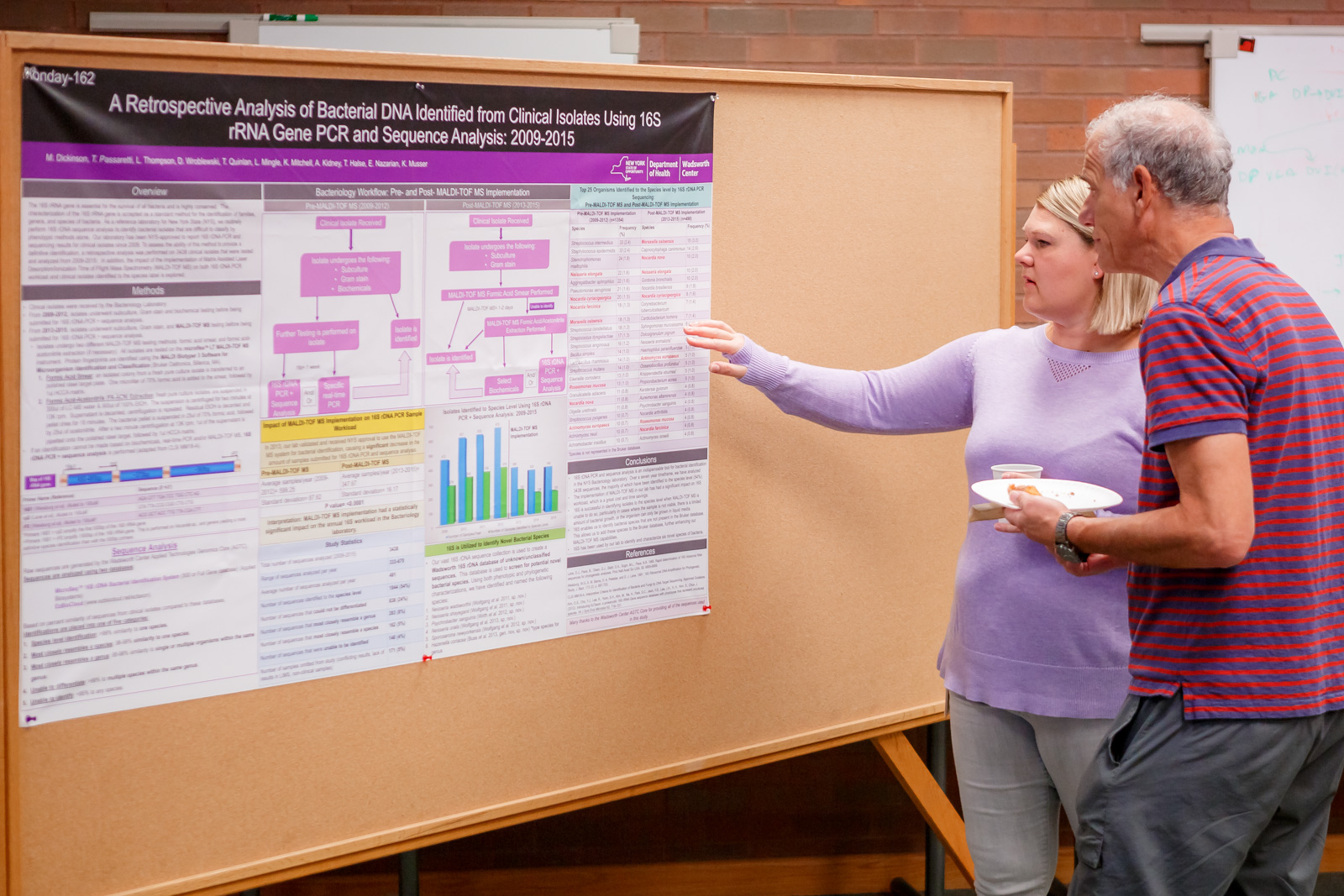 In a retrospective study, the Bacteriology Laboratory sequenced the 16s rDNA of 3,438 isolates obtained from 2009 – 2015. Results confirm that the method is extremely useful for classification.
In a retrospective study, the Bacteriology Laboratory sequenced the 16s rDNA of 3,438 isolates obtained from 2009 – 2015. Results confirm that the method is extremely useful for classification.
With increasing sample load comes the need for faster, more high-throughput methods. The Biodefense Laboratory demonstrated the utility of a CDC-developed Matrix-Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF/MS) method to identify botulinum neurotoxin and ricin toxin.
The Bloodborne Viruses Laboratory validated an alternative method of confirming HIV-1 positive results from dried blood spots. The APTIMA® HIV-1 RNA Qualitative assay is capable of obtaining accurate results on aged and heat treated samples.
HIV-2 RNA levels are so low in the infected patient that failure to detect HIV-2 RNA doesn’t necessarily constitute a negative diagnosis. The Bloodborne Virus Laboratory used droplet digital PCR to detect HIV-2 DNA and correlate DNA and RNA levels with diagnosis and treatment.
Real time RT-PCR confirmed the diagnosis of congenital rubella in two infants in NYS, a rarely seen syndrome in the US in more than a decade. Using a CDC-developed method, the Virology Laboratory further genotyped the isolates, matching them to those circulating in the country where the mothers were originally infected.
 The Virology Laboratory also demonstrated the use of self-avoiding molecular recognition systems (SAMRS) primers for improved specificity and sensitivity in multiplexed microarrays.
The Virology Laboratory also demonstrated the use of self-avoiding molecular recognition systems (SAMRS) primers for improved specificity and sensitivity in multiplexed microarrays.
Studies on influenza negative specimens from New York State college students with influenza like illness showed that human adenovirus was frequently the cause, demonstrating the importance of broader respiratory virus testing in order to avoid inappropriate use of influenza antivirals.
In a study of 942 influenza-negative specimens from the 2013-2014 influenza season, the CDC-designed Taqman Array Card (TAC) for the detection of 22 causes of respiratory disease was found to be an excellent surveillance tool.
The Laboratory of Clinical Microbiology is a partnership between the mycology, bacteriology, virology, blood borne virus, parasitology, mycobacteriology, rabies, arbovirology and diagnostic immunology laboratories. Seminars are open to the entire Department of Health community who are welcomed to share information and ideas for the development of assays to improve the health of New York State residents.