Nilesh Banavali, PhD
Infectious agents that cause human disease are sustained by a diverse range of their macromolecules performing specific functions. To understand disease mechanisms and develop therapeutics, visualizing detailed three-dimensional structures of such macromolecules is an important first step. This information needs to be connected to an understanding of the function of these macromolecules. Such a connection can be made by a description of the underlying free energy landscapes governing the activity of these macromolecules. The primary goals of our research are to use cryo-electron microscopy and computational structural biology techniques to characterize the detailed atomic structure of such macromolecular assemblies, understand the free energy landscapes of their activity, and translate this knowledge to structure-based discovery and optimization of therapeutic agents targeting their function. We typically validate our findings by additional experimental data through collaborations with other Wadsworth Center and external research groups. Our research is focused on the following areas:
Ribosome hibernation in human pathogens
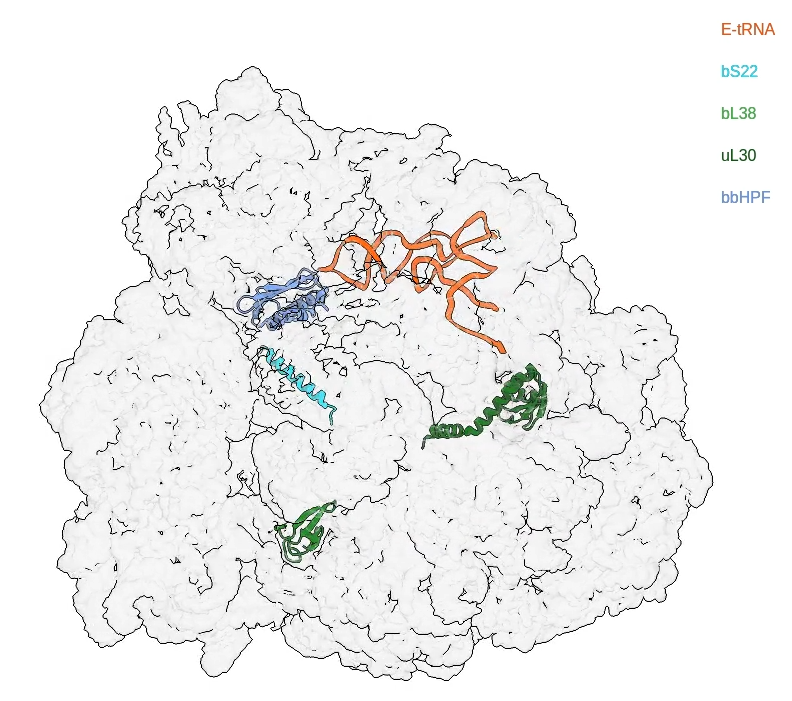
Bacterial cells consume a lot of their resources to synthesize proteins using their ribosomes. When conditions are poor for growth, they reduce protein synthesis using ribosome hibernation mechanisms that may also make them less susceptible to antibiotics. In a collaboration with the Agrawal and Lin groups at Wadsworth Center, we recently solved a 2.9 Å resolution structure of the hibernating 70S ribosome in Borrelia burgdorferi (Bbu), the spirochete bacteria causing Lyme disease (Sharma et al. 2023). The new discoveries in this work included the hibernation promoting factor protein bound to the decoding center of the 30S subunit in a location overlapping with the doxycycline binding site, two ribosomal proteins not previously known to be present in Bbu (bS22 and bL38), and a novel longer ribosomal uL30 protein, which plays the role of two bacterial ribosomal proteins – uL30 and bL37. This longer uL30 protein also exemplifies how the protein content of mammalian mitochondrial ribosomes might have increased during their evolution from a prokaryotic ribosome precursor. In a collaboration with the Agrawal and Ojha groups at Wadsworth Center, a 2.9 Å resolution structure of an alternative hibernating 70S ribosome in Mycobacterium smegmatis (Msm) that is produced in zinc-deficient growth conditions was also solved. We aim to use such structural and biochemical approaches to characterize ribosome hibernation and antibiotic resistance mechanisms in bacterial and fungal pathogens.
Structure-based therapeutic design
Half of the antibiotics in therapeutic use today are molecules that target ribosome function, indicating that the ribosome is one of the most important infectious disease therapeutic targets. Recent advances in machine learning-based computational prediction techniques have enabled accurate structure prediction for biological macromolecules. Complementation of these computational techniques with cryo-electron microscopy has the potential to greatly accelerate therapeutic discovery. We aim to combine our structural data with computational strategies to make structure-based improvements of known antibiotics that target bacterial ribosomes. This work is being pursued to improve antibiotics that target Lyme disease in collaboration with the Agrawal and Lin groups at Wadsworth Center and the Wang group at the University at Albany. We have previously demonstrated success in the use of such techniques to identify potential antiviral molecules against flaviviruses such as Zika virus, West Nile Virus, and Dengue virus in collaboration with the Li and Kramer groups at the Wadsworth Center (Chen et al. 2013, Li et al. 2017)
Free energy landscapes and macromolecular function
We have developed molecular simulation and free energy determination techniques to clarify multiple fundamental biochemical mechanisms starting from just structural models (Banavali and MacKerell 2002, Banavali and Roux 2005). Base pairing, or its reverse (base pair separation), is the main molecular mechanism involved in the three primary processes in the central dogma of biology: replication, transcription, and translation. We delineated the underlying free energy landscapes of the strand slippage mechanism by which a single base insertion or deletion (indel) mutation can occur in DNA strand extension and identified the possibility of specific non-canonical interactions providing a direct mechanism for sequence dependence of indel mutations (Banavali 2013, Manjari et al. 2014). We developed a strategy called Restrained Geometries and Topology Switching (RGATS) to obtain multiple iterative atomic-detail trajectories for biochemical reactions using standard macromolecular simulation methods (Manjari and Banavali 2018). The strategy is orders of magnitude faster than standard Quantum Mechanics/Molecular Mechanics (QM/MM) approaches. Its ability to rapidly explore different postulated mechanisms in atomic detail is ideally suited for computational biochemists to use as a first step before QM/MM calculations, or for experimentalists to identify mutations that could modulate specific steps of the reactions. The strategy has been successfully applied to model multiple functional mechanisms: (a) Proton transfer in an ammonia transporter, (b) nucleic acid strand extension in DNA polymerases, (c) autocatalytic cholesterol modification of hedgehog proteins (Banavali 2020).
Our long-range goals are to apply an optimal combination of structural, biochemical, and computational techniques to fully understand functional cellular machinery and to leverage that information to develop the next generation of therapeutics to address the perennial problems of emerging drug resistance.