Distributive Conjugal Transfer
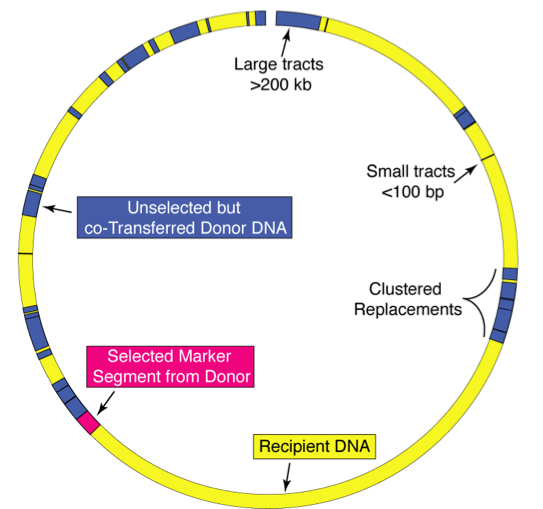
We have described a novel DNA transfer system in M. smegmatis that involves the transfer of chromosomal DNA between defined donor and recipient strains. The transconjugant genomes are a mosaic blend of the parental genomes and so we call the process distributive conjugal transfer (DCT). DCT is an entirely novel mechanism of horizontal gene transfer that has enormous potential to remodel the genome and facilitate rapid evolution. Our goal is to determine the mechanism of DCT and its prevalence among mycobacteria.
Protein Secretion
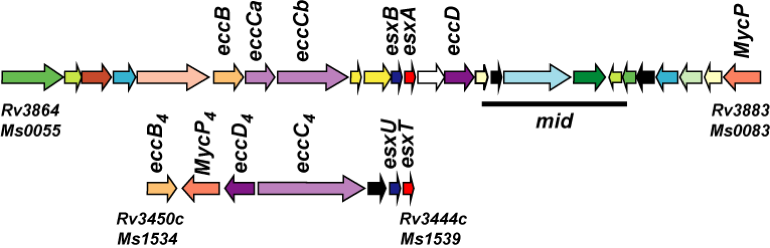
We have shown that the Type VII secretion systems, which are almost exclusively found in mycobacteria, play a major role in DCT. Specifically, the recipient must encode active ESX-1 and ESX-4 secretion systems for DCT. These secretion systems are of considerable interest as they are highly conserved and many have been shown to play a major role in pathogenesis. DCT provides a simple and efficient assay for ESX function that we have exploited to genetically characterize the proteins and mechanism of secretion. Our current goal is to determine the roles of these two ESX systems in the same biological process of DCT.
Cell-Cell Communication
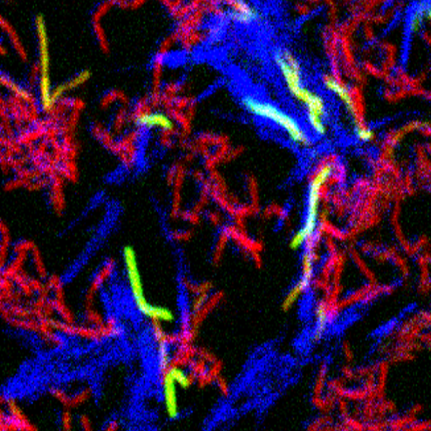
We have recently shown that mycobacteria communicate during DCT. When donor and recipient cells are co-cultured, the esx4 gene locus is highly induced. This induction occurs only in the recipient cells and it requires direct donor-recipient cell contact. The recipient response is also dependent on encoding a functional ESX-1 apparatus suggesting that ESX-1 acts upstream of ESX4 in the signaling pathway for DCT. In these experiments, we also identified hundreds of other transcripts that were only induced on co-culture indicating that intercellular communication is an important facet of mycobacterial growth. Our current goals are to determine; (i) the signaling pathway for ESX-4 expression, (ii) the contributions of the donor cell required for communication and esx4 induction, (iii) the significance of the many other contact-dependent transcripts and, (iv) the prevalence of cell-cell communication in other mycobacterial species.
Gene expression and architecture
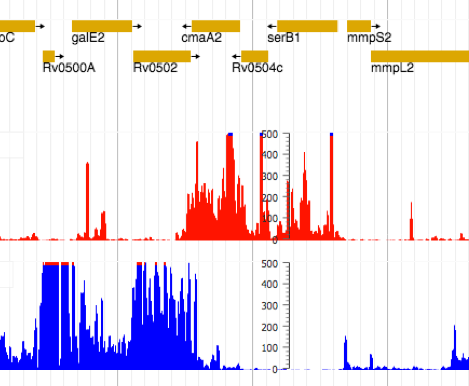
In collaboration with our Wadsworth colleague, Dr. Joe Wade, we have performed extensive transcriptional and translational profiling analyses on M. smegmatis and M. tuberculosis. These data are freely available to the scientific community to identify their gene(s) of interest.
The ability to precisely map the 5’ends of mRNAs and the start sites of translation initiation highlighted two unusual features of gene architecture in mycobacteria. First, mycobacteria encode many leaderless transcripts (they lack a 5’UTR, including the Shine-Dalgarno) sequence. Second, they encode hundreds of small proteins, or sproteins, (less than 50 amino acids). Thus, many genes have been mis-annotated or missed in current mycobacterial annotations. Our current work is focused on: (i) re-annotating the genome sequences based on our experimental data, (ii) determining the mechanism of leaderless translation, and (iii) identifying the functions of sproteins.
Mycobacterial Systems Resource
In collaboration with the laboratories of Drs. Joe Wade, Graham Hatfull (University of Pittsburgh) and Eric Rubin and Sarah Fortune (Harvard), we have begun to establish an M. smegmatis based resource. The resource is based on 1153 M. smegmatis proteins that have >50% amino acid identity with orthologues encoded in M. tuberculosis, M. leprae, M. ulcerans and M. avium. This level of conservation across all species ensures that these proteins mediate fundamental mycobacterial processes, and that any genetic, phenotypic and bioinformatic data generated from their analysis will be applicable to all species of mycobacteria.
Our goal is to create: (i) a physical resource of M. smegmatis strains and plasmid derivatives of the 1153 genes; (ii) a phenotypic resource based on the analysis of these strains; and (ii), a bioinformatic resource providing precise genome annotations and experimental details of the physical resource. Data integrated from all three resources will be available online for all researchers and will provide an informed basis from which to design experimental approaches in the less tractable pathogens.
To date the physical resource consists of strains and plasmids containing derivatives of the 1153 genes. We have created; (i) 601 knock-out strains of non-essential genes and (ii), 1118 C-terminal gene fusions to the gene encoding the fluorescent protein Dendra. These two libraries are complete and ready for depositing at BEI.