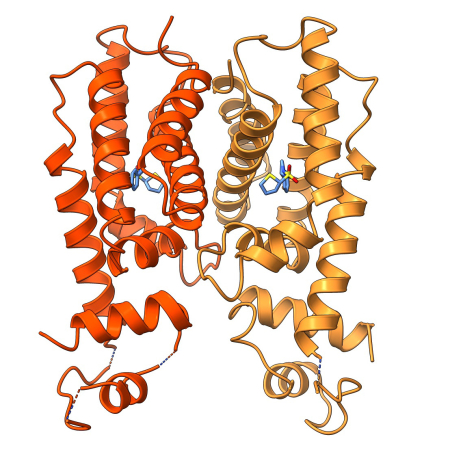
Research in the Paczkowski laboratory focuses on the mechanisms of regulation of quorum sensing (QS) in Gram-negative bacteria, such as Pseudomonas aeruginosa, Vibrio cholerae, Aeromonas hydrophila, and Chromobacterium violaceum. QS is a process of bacterial cell-cell communication that controls virulence and biofilm formation in many bacterial species. QS relies on the production, accumulation, detection, and population-wide response to extracellular signal molecules called autoinducers (AI). QS allows bacteria to synchronously alter gene expression patterns that underpin collective behaviors. Using QS as a model will allow us to understand long-standing questions in the field related to signal recognition preferences in single species and multi-species environments and the regulation of the interrelated signaling cascade that exists to coordinate behavior. Learning how bacteria correctly interpret these blends of AIs and elicit appropriate gene expression responses is essential to understand how bacteria communicate, and, more globally, to understand how all organisms decode environmental stimuli. QS presents a unique opportunity to understand these longstanding questions in biology.
Specific projects include:
- Determining the mechanisms used by regulators to control QS receptor function and recognition of DNA.
- Determining the mechanism of AI recognition for QS receptors.
- Screening small molecule libraries for inhibitors of QS receptors.
- Determining the role of QS in shaping microbial community architecture.
We use a combination of bacterial genetics, biochemistry, structural biology, and chemical biology to understand how bacteria respond appropriately to signals in a complex environment.
Join us!